Module: Microbial Systematics Course
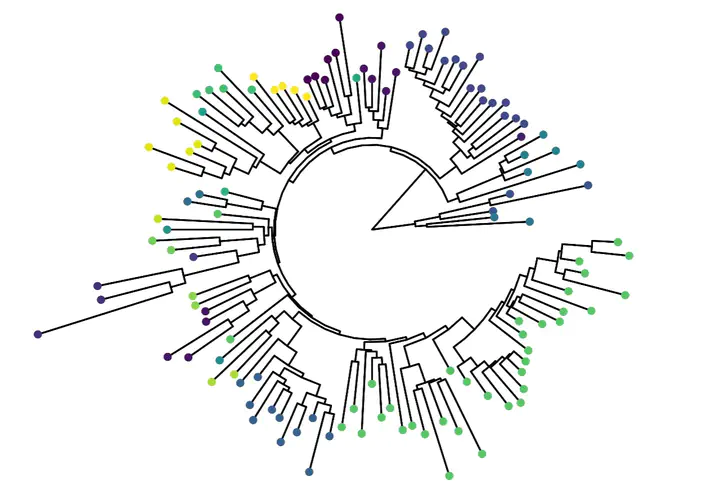 Phylogenetic Tree
Phylogenetic TreeModule: Computational Microbiology in the Microbial Systematics Course
Welcome to Computational Microbiology, a core component of the Microbial Systematics course. This module was developed to give students practical experience in analyzing microbial diversity and understanding the principles of taxonomy and classification using real-world data.
This project was created during my time as a lab assistant in the Microbial Systematics course at Institut Teknologi Sepuluh Nopember (ITS) Surabaya.
Module Objectives
Students will explore microbial samples from various environments, ranging from soil and water to more specialized ecosystems,and apply computational tools in R to:
- Understand microbial taxonomy and classification
- Retrieve and align genetic sequences
- Construct and interpret phylogenetic trees
What You’ll Learn
Dataset Preparation
Select diverse microbial species from various ecological niches for phylogenetic analysis.Genetic Data Retrieval
Access and retrieve DNA sequence data from GenBank for selected microbial taxa.Sequence Alignment
Perform multiple sequence alignment using bioinformatics tools to identify conserved regions and ensure accurate comparisons.Phylogenetic Tree Construction
Build evolutionary trees using methods like neighbor-joining or maximum likelihood, and learn how different approaches impact the resulting topology.
Note: This module is currently available in Bahasa Indonesia.
Explore the Module
Want to Collaborate?
I’m always open to discussing education projects, bioinformatics modules, or teaching collaborations. Feel free to get in touch!