R tutorial series: Analyzing bacterial growth from OD600 data
Bacterial growth analysis is a fundamental task in microbiology, allowing researchers to monitor the proliferation of bacteria over time. Optical Density at 600 nm (OD600) is a commonly used method to measure this growth. In this tutorial, we’ll walk through the process of analyzing bacterial growth data using R. We’ll cover data loading, cleaning, visualization, and calculating key growth metrics such as the growth rate and doubling time. This is just basic things for a new undergrad biology student and a nice way to start learning microbiology.
Libraries we need
We start by defining the libraries that will be used in our analysis. Let’s get our R environment ready before we dive into the data. We’ll need to make sure we have the right packages for everything from cleaning up data to making it look good.
libs <- c(
"readr", "dplyr", "lubridate",
"ggplot2", "ggsci", "plotly"
)
readr for efficient handling of data files, especially CSVs; dplyr for data manipulation and transformation; lubridate for working with dates and times; ggplot2 for creating static visualizations; ggsci for enhancing plot aesthetics with scientific color palettes; and plotly for interactive plot creation based on ggplot2 outputs.
# Install missing libraries
installed_libs <- libs %in% rownames(installed.packages())
if (any(installed_libs == FALSE)) {
install.packages(libs[!installed_libs])
}
# Load libraries
invisible(lapply(libs, library, character.only = TRUE))
lapply(): This function loads each library in the list. The character.only = TRUE argument allows the function to use the names of the libraries stored in the libs vector.
invisible(): This function suppresses the output that library() normally generates when loading a package, keeping the console output clean.
Load and preprocess the data
Once our libraries are ready, we can load our OD600 data from a CSV file, you can also use excel and makes sure the table looks exactly the same. You can download the data here. This data is my personal data that I got during my training in The University of Vienna. I worked with Vibrio as a bacterial model in marine broth liquid medium.
df <- read_csv("od_data2.csv") %>%
mutate(Sample = sub("_.*", "", Sample_name),
Time = sub(".*_", "", Sample_name),
Result = Result - 0.001,
Result_log = log(Result)) %>%
select(Sample, Time, Result, Result_log) %>%
mutate(Time = hm(Time))
Data preprocessing involves loading the CSV data into a dataframe using read_csv. We extract sample name and time from the Sample_name column using mutate and regular expressions. The OD600 readings are adjusted by subtracting a blank value and log-transformed for subsequent analysis. Note that you need to adjust the value based on the blank value measurement, it is important to assure we get the real OD value. Finally, the Time column is converted into a time object using the lubridate package’s hm function.
Plot the growth curve
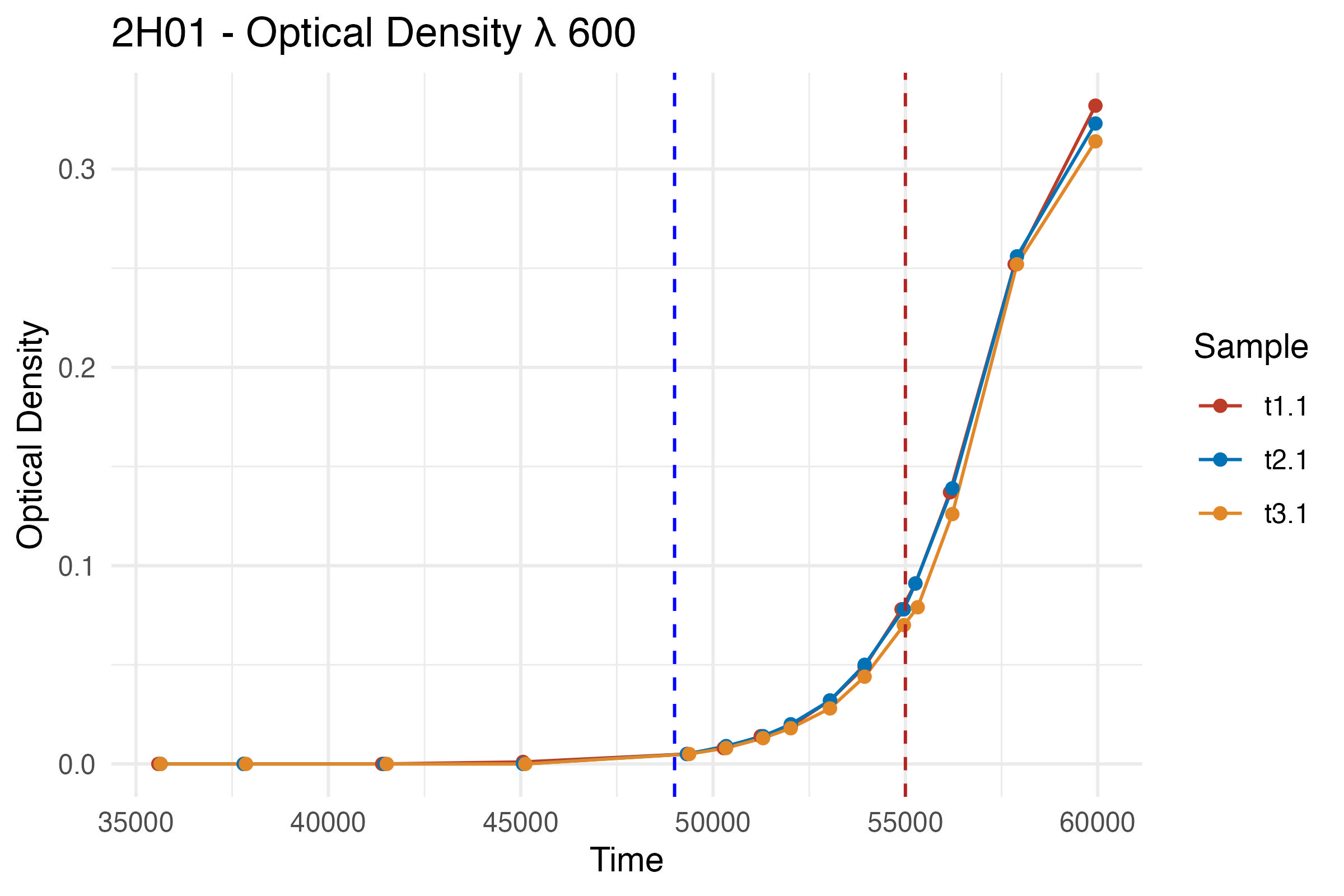
Now that our data is ready, let’s visualize the bacterial growth over time using ggplot2. Visualizing the data helps in understanding the growth pattern and identifying phases like the lag and exponential growth phases.
p <- ggplot(df, aes(x = Time, y = Result, color = Sample)) +
geom_line() +
geom_point() +
labs(title = "2H01 - Optical Density λ 600",
x = "Time",
y = "Optical Density") +
scale_color_nejm() +
theme_minimal() +
geom_vline(xintercept = 49000, linetype = "dashed", color = "blue") +
geom_vline(xintercept = 55000, linetype = "dashed", color = "firebrick")
p
# Save the plot
ggsave("growthcurve.jpeg", p, width = 6, height = 4, units = "in", dpi = 400)
Interactive plot
ggplotly(p)
Plot the log-transformed OD600 data
ggplot(df, aes(x = Time, y = Result_log, color = Sample)) +
geom_line() +
geom_point() +
labs(title = "2H01 - Optical Density λ 600 (Log Scale)",
x = "Time",
y = "Log(Optical Density)") +
scale_color_nejm() +
theme_minimal() +
geom_vline(xintercept = 49000, linetype = "dashed", color = "blue") +
geom_vline(xintercept = 55000, linetype = "dashed", color = "firebrick")
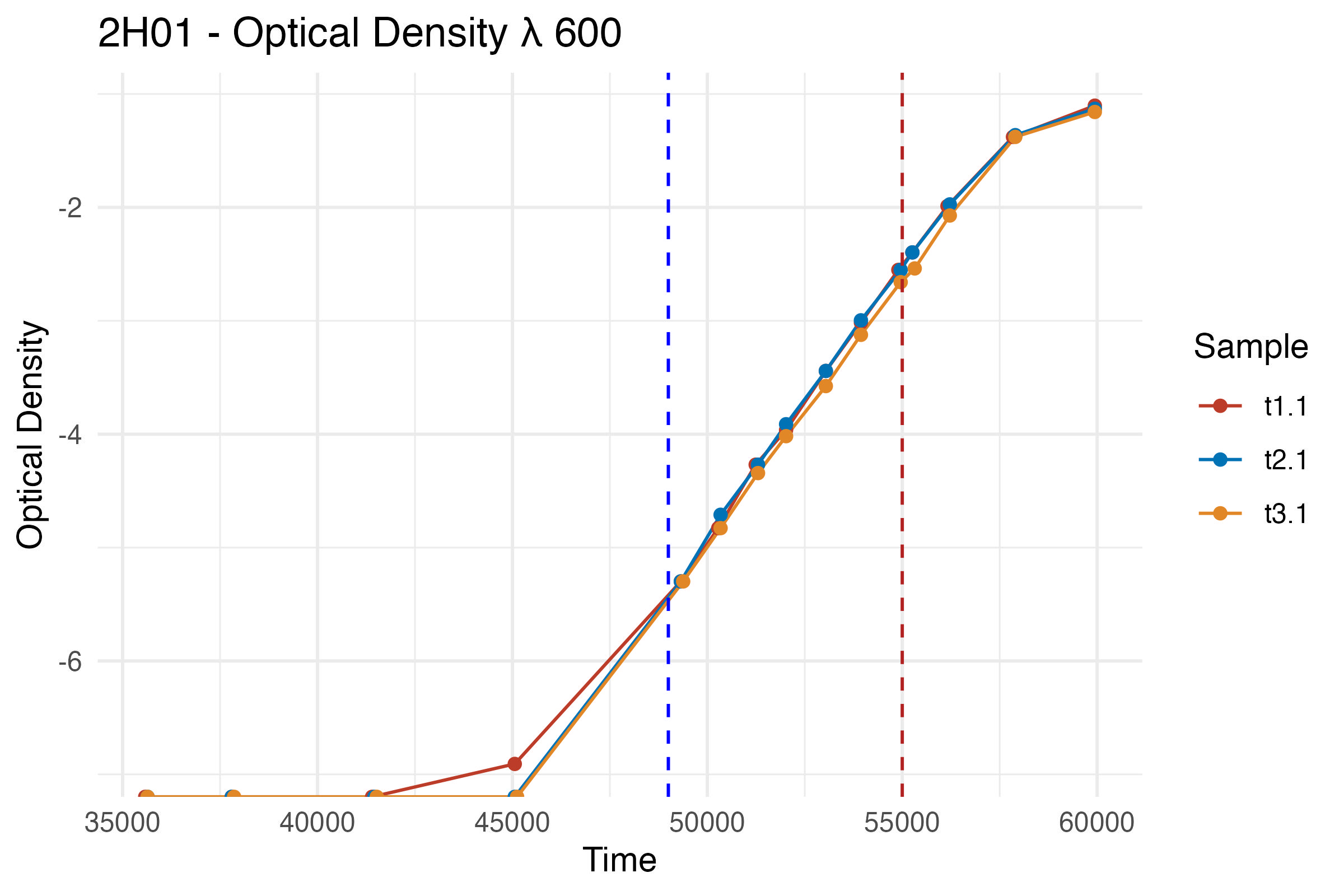
# Define lag and exponential phase endpoints
lag_end <- hm("13:00") # Assumption based on the log-transformed curve
exp_end <- hm("15:30")
# Filter data for the exponential phase
exp_phase_data <- df %>%
filter(Time > lag_end & Time < exp_end)
Fit an exponential model
Fits a linear model to the log-transformed OD600 data, which is equivalent to fitting an exponential model to the original data.
model <- lm(Result_log ~ as.numeric(Time), data = exp_phase_data)
summary(model)
Calculate the growth rate and doubling time
To understand how fast the bacteria are growing, we calculate the growth rate and the doubling time during the exponential phase. In batch culture, the growth of bacteria can be analyzed using exponential growth models. Here’s how you can calculate the growth rate and doubling time.
Growth Rate
The growth rate can be expressed as a differential equation below
$$ \frac{dN}{dt} = kN $$
where kN is the rate of change in population size, N, at a given moment in time, t. In R, we can just extract the second coefficient of the model because if we derivate the function, we’ll get second coefficient left.
# Growth rate per hour
growth_rate_per_hour <- coef(model)[2] * 3600
growth_rate_per_hour
Doubling Time
The doubling time (td) is the time required for the bacterial population to double. It is derived from the growth rate using the formula:
$$ t_d = \frac{\ln(2)}{kN} $$
doubling_time <- log(2) / coef(model)[2]
doubling_time/60 # In minutes period
If you follow the steps above correctly, you will get 24.1566 minutes as a doubling time.
Remarks
By following these steps, we’ve successfully analyzed bacterial growth data, visualized the growth curve, and calculated important metrics like the growth rate and doubling time. This process is essential in microbiology for understanding how bacteria respond to different conditions over time.
Buy me a coffee See you in the next post and have a beautiful day!